Abstract
Background: Diffuse large B-cell lymphoma (DLBCL) is a highly heterogeneous disease, and the high-throughput sequencing has facilitated our understanding of genetic aberrations in DLBCL. The proviral integration site for Moloney murine leukemia virus 1 (PIM1), which encodes serine/threonine protein kinase, is identified as a target of aberrant somatic hypermutation in DLBCL and involved in tumorigenesis in hematopoietic malignancies. However, there are few studies focused on its genetic alterations, molecular profiles, drug responses and clinical significance.
Materials and methods: We integrated targeted sequencing and transcriptome analysis to explore the pathogenic role of PIM1 mutations and as a personalized therapeutic target in PIM1-mutated DLBCL patients.
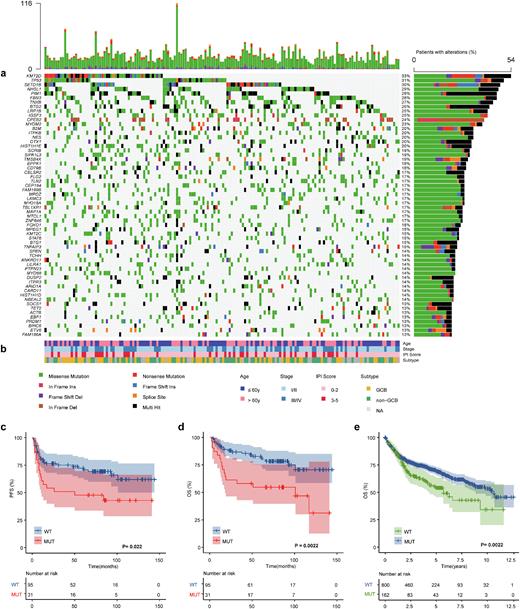
Results: We found PIM1 to be mutated in 28.4% patients with DLBCL, with 164 genetic alterations. Variant classifications showed that missense mutations occurred most frequently (84.1%); almost half of them (48.7%) are predicted to be deleterious (SIFT score<0.05). Besides, the C>T transition was the predominant type (54.4%). Of the 46 mutant patients, all samples harbored nonsynonymous alterations, with more than 3 mutations detected in a single sample from half of the patients. We observed exon 4 to most often be mutated, and 57% (84/164) of mutations are located in the serine/threonine protein kinase domain. Comutation and mutual exclusivity analysis identified 72 statistically significant interaction pair genes, of which PIM1 mutations significantly co-occurred with SETD1B (p<0.001), CD79B (p=0.001) and MYD88 (p<0.001) but not with SPEN mutations (p=0.024). We also found that patients with PIM1 mutations had higher mutation frequencies in PRDM1 (p<0.001) and CD79B (p=0.001) involved in the NF-κB pathway and B cell receptor (BCR) pathway.
Compared with wild-type patients, those with mutations had significantly higher IPI scores (p=0.031), especially in the high-risk subgroup (17.4% vs. 4.3%), and were more likely to relapse (50% vs. 32%, p=0.031); there was a trend toward a higher proportion in the non-GCB subtype (52% vs. 39%). In particular, patients harboring PIM1 mutations more frequently had testis and/or CNS involvement (73% (8/11) vs. 25% (38/151), p=0.001). Progression-free survival (PFS) and overall survival (OS) were significantly shorter in the mutation group than in the wild-type group (PFS, p=0.022; OS, p=0.0022), which was confirmed in the external validation cohort (p=0.0022). In multivariate Cox analysis, PIM1 mutation remained an independent unfavorable prognostic factor (p=0.004).
Transcriptome sequencing revealed that PIM1 mutation led to a significantly higher level of gene expression (p<0.001). Furthermore, several upregulated genes (n=175) involved in the immune response (IGLC6, IGLJ6, CLEC4C), posttranslational modification (ADPRHL1, NEURL1), nuclear RNA export (NXF3), carcinogenesis (WIF1, WNT9A) and transcription factors (HMX3,ZNF320) were enriched in patients with PIM1 mutation. KEGG analysis revealed disorder of the tumor microenvironment (e.g., cytokine-cytokine receptor interaction, chemokine signaling, TNF signaling), JAK-STAT and NF-κB pathways in patients with PIM1 mutation, which were consistent with protein-protein interaction (PPI) network analysis.
A novel PIM1 mutation-related gene signature further indicated distinct phenotypes and served as a promising biomarker for risk stratification. Patients with high-risk score had higher response to TGFβ receptor inhibitors (SB525334, SB505124, p<0.0001), Lenalidomide (p=0.041), NF-κB inhibitors (Parthenolide, TPCA-1, p<0.0001) and JAK inhibitors (Ruxolitinib, TG101348, p=0.014). Our findings suggest that the novel signature not only improves prognostic stratification but also provides personalized therapeutic decisions for patients with high risk.
Conclusions: Our findings reveal that PIM1 mutation is involved in the pathogenesis of DLBCL, suggesting that detection of PIM1 mutations with incorporation of our PIM1 mutation-related gene signature will be helpful for identifying DLBCL patients at high risk of progression and might provide predictive information for the design of personalized therapeutic strategies.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal